Simple evolutionary study may predict path of Ebola outbreaks
 Ebola is one of my favorite pathogens. With the reputation it has, many people assume it’s killed many more worldwide than it actually has. People hear of Ebola and all kinds of grotesque images come to mind: organs “liquefying” (doesn’t really happen quite like that); bleeding from every orifice (okay, that one can be on-target); the victims dying a horrible death from a virus with an incredibly high mortality rate. There are four known subtypes of Ebola, named for their place of isolation: Ebola Reston, Ivory Coast, Sudan, and Zaire. Together with their cousin, the Marburg virus, they make up the family of viruses known as filoviruses.
Ebola is one of my favorite pathogens. With the reputation it has, many people assume it’s killed many more worldwide than it actually has. People hear of Ebola and all kinds of grotesque images come to mind: organs “liquefying” (doesn’t really happen quite like that); bleeding from every orifice (okay, that one can be on-target); the victims dying a horrible death from a virus with an incredibly high mortality rate. There are four known subtypes of Ebola, named for their place of isolation: Ebola Reston, Ivory Coast, Sudan, and Zaire. Together with their cousin, the Marburg virus, they make up the family of viruses known as filoviruses.Marburg was the first of these to be recognized, causing an outbreak in Germany (caused by infected African research monkeys) in 1967. The Ebola Zaire strain (EBO-Z) and the Ebola Sudan strain (EBO-S) surfaced at almost the same time in 1976. The outbreak in Zaire resulted in 319 cases (90% mortality), while in Sudan, 284 cases were identified (53% mortality rate). EBO-Z then wasn’t seen for almost 20 years, re-surfacing in Gabon in 1994, and once again in Zaire (Democratic Republic of Congo) in 1995. Another EBO-Z outbreak occurred in 2001-2 in Gabon and The Republic of Congo, causing about 120 cases, 79% of them fatal. Overall, less than 2000 known human infections and 1100 deaths have resulted from Ebola since its discovery in 1976. That’s an average of 38 deaths worldwide per year over the last 29 years. Compare that to a virus such as influenza, which kills 36,000 every year in the United States alone. Or even a fairly common microbe like E. coli, which causes thousands of deaths each year due to bacterial sepsis. Worse, none of these even come close to malaria, which causes over 200 deaths worldwide every hour. The numbers make it clear that, as far as mortality goes, Ebola is small potatoes—we have more to fear from our hamburger than from this exotic African virus. Yet, the Ebola mystique lingers.
One reason for this is because, despite years of study and millions of dollars spent, Ebola remains mysterious. Mystery and unfamiliarity breed fear. (Dramatic Dustin Hoffman movies tend to assist in that effort as well). We have no idea what the reservoir of the virus is in nature. We don’t know how it moves from that reservoir into the human (or great ape) population. We don’t know the extent of its distribution in nature—-the Reston strain originated in a shipment of monkeys from the Philippines, so filoviruses must be present outside of Africa. Finally, we don’t know where it might hit next.
A new study by Walsh et al. tries to address that final concern. They ask the question, has EBO-Z long been present in the region where outbreaks have occurred? Or has the virus spread recently to the outbreak area? To begin answering this question, they overview the relationship between viral phylogeny and host-virus dynamics. They explain that it is the tendency of viruses which have been maintained within a single host population to have a high genetic diversity. An example they give of this is Hepatitis C, which has a number of different genotypes with worldwide distribution. However, the greatest viral diversity is found in populations where the virus has long been endemic. Alternatively, more limited genetic diversity is a characteristic of viruses that are either spreading, or undergoing positive selection. Influenza in humans is an example of this. The authors of the current study used this information, along with sequence data from Ebola viruses isolated during outbreaks occurring between 1976 and 2002 (see figure 1 below), in their study.
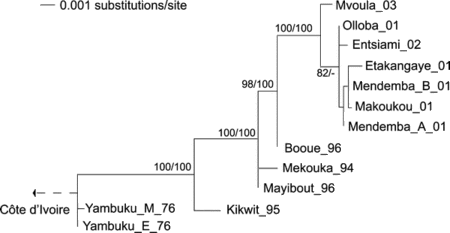
Figure 1 ML Tree of Full-Length (> 2,000 bp) ZEBOV-GP SequencesFirst, they tested whether the turnover of Ebola viral genotypes was due to positive selection for new variants. A classic way to examine this is to look at the ratio of non-synonymous to synonymous substitutions in the genome. Recall that because of the degeneracy of our genetic code, not every mutation in the genetic sequence will result in a change in the amino acid sequence of the encoded protein. (synonymous changes=no change in amino acid; non-synonymous changes=change seen in amino acid). So, by measuring the ratio of these changes, they can infer if there is directional selection occurring. Long story short, they concluded that this may have had a small effect, but it does not seem to be a major determinant in the emergence of new viruses.
Tree was found in Paup* and rooted in a separate analysis using ICEBOV as an out-group. The latter analysis excluded a 576-bp variable region for which alignment with ZEBOV was uncertain. Numbers next to branches indicate percent support based on 1,000 bootstrap replicates and posterior probabilities obtained in a molecular clock-based analysis in program BEAST (only values > 70% are shown).
They next looked at the geography of the outbreaks, and its correlation to new virus genotypes. For example, between 2001-2004 the spread of the virus eastward was consistent (46.1 km per year); this remained similar even when they added in earlier 1996 outbreaks. They argue this suggests a single spread eastward. Working backward to find the origin of the ancestral strain, their data suggested a site northwest of Yambuku, Zaire in the year 1973. They also were able to corroborate with their molecular data an observation that had previously been made: that the virus front seemed to shift following the 1996 outbreak in Booué, Gabon. Additionally, the authors got a test of their predictions as their paper was being reviewed. A May 2005 outbreak in Etoumbi village (also in Gabon) fit nicely into their prediction model.
What does all this mean? For one, it lends support to the hypothesis that Ebola is truly an emerging disease, in every sense of the word. The Zaire strain appears to be spreading out from Yambuku; all EBO-Z to date are descendants of the original Yambuku strain. Positive selection does not appear to be a determining factor, though it may play a small role. Finally, increasing genetic divergence was found to correlate with increasing geographic distance.
There are certainly limitations to the study. Most importantly, small sample sizes, such as they had in this study, limit the accuracy of their model and the predictive power of their conclusions. Still, this elegant study based on simple evolutionary principles also has real-world applications. As mentioned earlier, no virus reservoir has ever been identified. Using knowledge of the spread of EBO-Z out of Yambuku, ecologists can search for candidate reservoir species that fit this distribution; a species whose range may have similarly expanded. It also suggests a new paradigm for control of the virus: focus on the spread of the “advancing wave.” This is an area that could also be targeted for an Ebola vaccine, when one is eventually produced.
Additionally, I mentioned above that there are 4 known distinct lineages of Ebola: Zaire, Sudan, Ivory Coast, and Reston. This study only focused on the most virulent; the Zaire strain. It doesn't answer the question of whether EBO-Z is spreading into a population where other filoviruses already exist, but EBO-Z is out-competing them, or if it's spreading into regions where no Ebola relatives are present. Just based on other epidemiologic data, I'd guess it's the former. Serologic studies in several regions of Africa have shown that many people possess antibodies to the Ebola virus, even though they report never having had a hemorrhagic fever (or experiencing an outbreak of Ebola-like illness in their village). It seems likely that filoviruses are present somewhere in the environment already, but these strains are probably low-virulence, and do not cause overt disease. The Reston strain already has been shown to cause asymptomatic infection with humans; there is no reason to think others like this cannot be out there as well. Are they losing an evolutionary battle with EBO-Z? If so, that suggests that we may see more frequent outbreaks in the future.
 Finally, there is another ominous prediction. If the rate of spread continues at its current pace, EBO-Z may hit populated areas outside of Odzala National Park in the coming year or two, putting greater numbers of not only humans, but also great apes, at risk. Other areas with large gorilla populations—already decimated by poaching and Ebola in Africa--could be reached within 3-6 years. The truly frightening conclusion is that the worst Ebola has to offer may be yet to come.
Finally, there is another ominous prediction. If the rate of spread continues at its current pace, EBO-Z may hit populated areas outside of Odzala National Park in the coming year or two, putting greater numbers of not only humans, but also great apes, at risk. Other areas with large gorilla populations—already decimated by poaching and Ebola in Africa--could be reached within 3-6 years. The truly frightening conclusion is that the worst Ebola has to offer may be yet to come.


